Quantitative biology of cancer metabolism
The Warburg effect
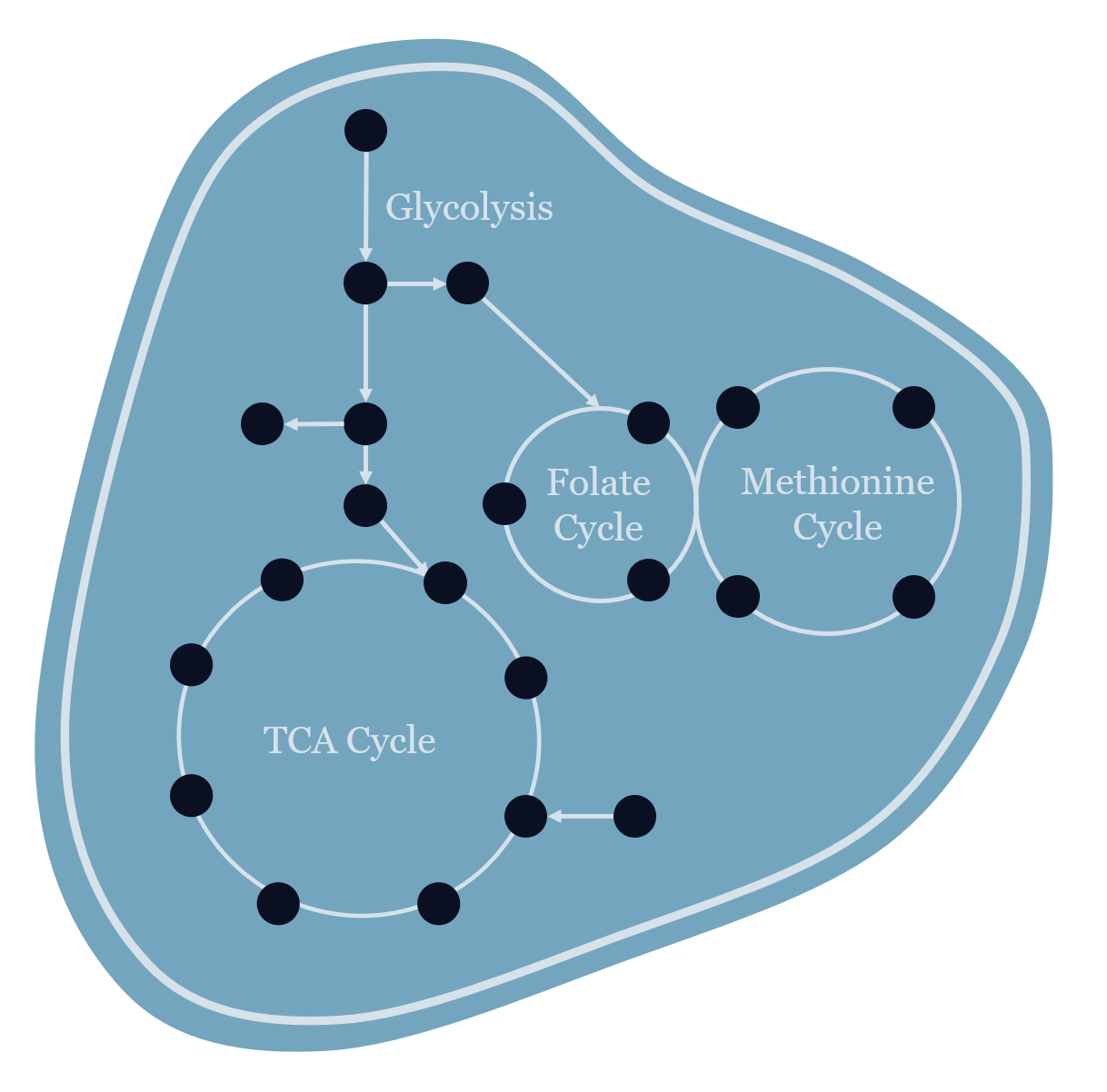
To investigate possible mechanisms that cause the Warburg Effect (WE) in cancer cells, we developed a flux balance model of glycolysis to demonstrate that the extent of WE in cancer cells is determined by cellular demands of redox balancing, energy and biomass (Dai et al, Biophysical Journal 2016). We then developed a more detailed ODE model for glycolysis and performed metabolic control analysis with this model to identify the enzyme GAPDH as the rate-limiting step of glycolysis in cells with higher WE. The prediction was validated by experiments in cancer cells and mouse xenograft tumor models which showed that KA, an inhibitor of GAPDH, selectively killed cancer cells with higher WE and reduced tumor growth in vivo (Liberti, Dai et al, Cell Metabolism 2017). These findings together identify WE as a metabolic signature predictive of tumor cells’ response to therapeutics targeting GAPDH. We also developed a genome-scale, multi-objective optimization model to study requirements of cancer cell metabolism and identify novel metabolic targets for cancer therapeutics (Dai et al, Cell Commun Signal 2019).
Methionine, epigenetics and cancer
Methionine is an essential amino acid with multi-faceted roles in regulating metabolism, epigenetics and health. We used the epigenetic mark histone H3 lysine 4 trimethylation (H3K4me3) as a model to study its responses to methionine restriction in cancer cells and liver of mice under normal and methionine-restricted conditions (Dai et al, Nature Communications 2018). Using quantitative metrics we developed, we found that changes in H3K4me3 width were strongly associated with cell type specific biological functions, cell identity-related transcription factor (TF) binding motifs, and changes in gene expression. These results demonstrate that changes in methionine metabolism lead to changes in H3K4me3 peak width that encode both gene expression dynamics and cell identity information.
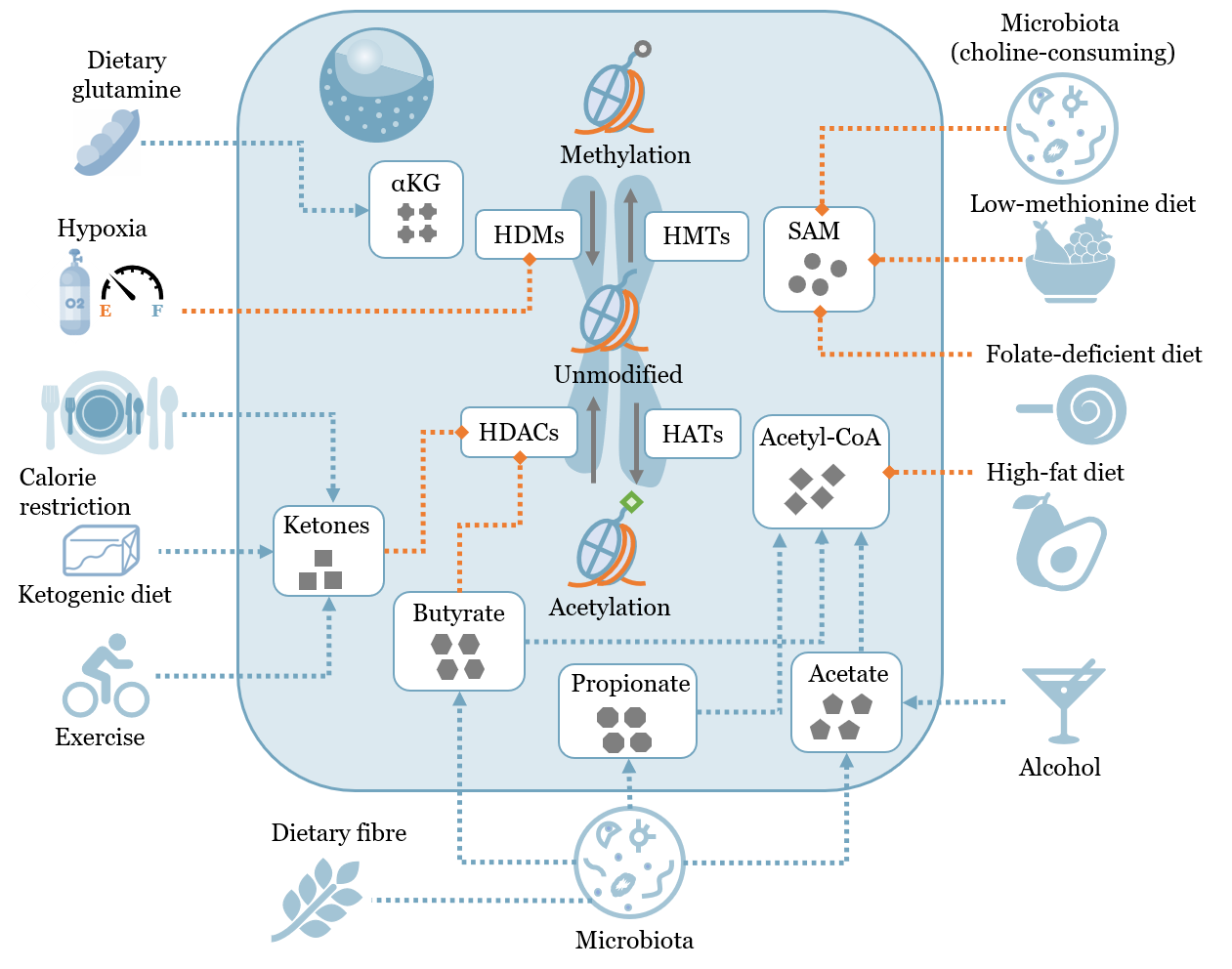
We also studied the effects of dietary methionine restriction on metabolism and cancer outcomes in mouse tumor models including patient-derived xenograft model and sarcoma model (Gao, Sanderson#, Dai# et al, Nature 2019). We applied computational approaches including singular value decomposition and genome-scale metabolic network analysis to show that dietary methionine restriction specifically targets one carbon metabolism in multiple tissues to mediate its anti-tumor effects, and that similar changes in plasma metabolite profiles can be achieved in humans through dietary methionine restriction.
Metabolic landscape of tumor microenvironment
To dissect the metabolic landscape of the tumor microenvironment at the single-cell resolution, we developed a computational pipeline and applied it to single-cell RNA-seq datasets for two human tumor types including melanoma and head and neck (Xiao, Dai* and Locasale*, Nature Communications 2019). We found that single tumor cells exhibit higher metabolic activity and variation compared to single non-malignant cells and bulk tumors, and identified mitochondrial metabolism as the essential contributor to metabolic heterogeneity in both malignant and non-malignant cells.